CMU and Columbia Researchers Magnify Hidden Biological Structures with MAGNIFIERS
The team from CMU and Columbia combined microscopy methods to better view proteins, lipids and more at the nanoscale
Media Inquiries
A research team from Carnegie Mellon University and Columbia University have combined two emerging imaging technologies to better view a wide range of biomolecules, including proteins, lipids and DNA, at the nanoscale. Their technique, which brings together expansion microscopy and stimulated Raman scattering microscopy, is detailed in Advanced Science(opens in new window).
Biomolecules are traditionally imaged using fluorescent microscopy, but that technique has its limitations. Fluorescent microscopy relies on fluorophore-carrying tags to bind to and label molecules of interest. These tags emit fluorescent light with a broad range of wavelengths; thus researchers can only use 3-4 fluorescent colors in the visible spectrum at a time to label molecules of interest.
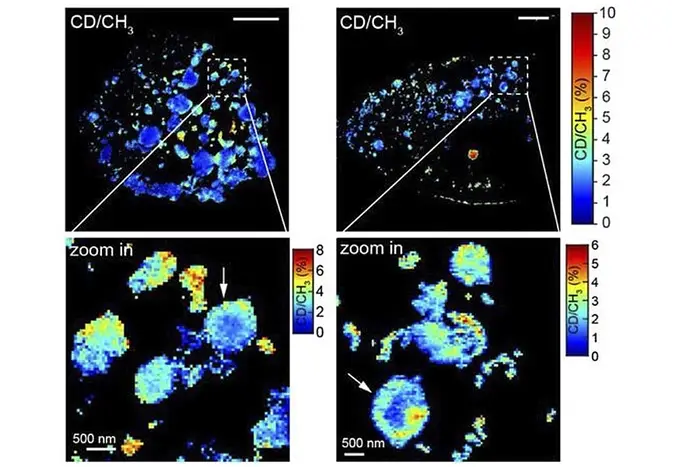
MAGNIFIERS captured newly synthesized Huntingtin aggregates (in red) with nanoscale resolution in the Huntington disease model.
Unlike fluorescent microscopy, stimulated Raman scattering microscopy (SRS) visualizes the chemical bonds of biomolecules by capturing their vibrational fingerprints. In this sense, SRS doesn't need labels to see the different types of biomolecules, or even different isotopes, within a sample. In addition, a rainbow of dyes with unique vibrational spectra can be used to image multiple targets. However, SRS has a diffraction limit of about 300 nanometers, making it unable to visualize many of the crucial nanoscale structures found in cells and tissue.
"Each type of molecule has its own vibrational fingerprint. SRS allows us to see the type of molecule we want by tuning in to the characteristic frequency of its vibrations. Something like switching between the radio stations," said Yongxin (Leon) Zhao(opens in new window), CMU's Eberly Family Associate Professor of Biological Sciences.
Zhao's lab has been developing new imaging tools based on expansion microscopy — a technique that addresses the problem of diffraction limits in a wide range of biological imaging. Expansion microscopy takes biological samples and transforms them into water-soluble hydrogels. The hydrogels can then be treated and made to expand to more than 100 times their original volume. The expanded samples can then be imaged using standard techniques.
"Just as SRS was able to surmount the limitations of fluorescence microscopy, expansion microscopy surmounts the limitations of SRS," said Zhao.
The Carnegie Mellon and Columbia researchers combined SRS and expansion microscopy to create Molecule Anchorable Gel-enabled Nanoscale Imaging of Fluorescence and stimulated Raman Scattering microscopy (MAGNIFIERS). Zhao's expansion microscopy technique was able to expand samples up to 7.2-fold, allowing them to use SRS to image smaller molecules and structures than they would be able to do without expansion.
In the recently published study, the research team showed that MAGNIFIERS could be used for high-resolution metabolic imaging of protein aggregates, like those created in conditions like Huntington's disease. They also showed that MAGNIFIERS could map the nanoscale location of eight different markers in brain tissue at one time.
The researchers plan to continue to develop the MAGNIFIERS technique to achieve higher resolution and higher throughput imaging for understanding the pathology of complex diseases, such as cancer and brain disorders.
Additional study co-authors include: Alexsandra Klimas, Brendan Gallagher, Zhangu Cheng, Feifei Fu, Piyumi Wijesekara and Xi Ren from Carnegie Mellon; and Yupeng Miao, Lixue Shi and Wei Min from Columbia.
This research was funded by the National Institutes of Health (DP2 OD025926-01, R01 GM128214, R01 GM132860, and R01 EB029523), Carnegie Mellon University, the DSF Charitable Foundation and U.S. Department of Defense (VR190139).
— Related Content —

Microscopy and VR Illuminate New Ways to Treat Diseases